HELP |
Retina is the specialized tissue involved in the light stimulus transduction. Despite its unique biology and the burden of genetic disease an accurate reference transcriptome is still lacking. Here we collected 50 high-quality post-mortem human retinas from donors and performed high-coverage RNA-sequencing analysis to yield a comprehensive reference transcriptome of the human retina.
The reference transcriptome includes 13,792 genes (download genelist) across 94,521 transcripts.
All the raw data are publicly available (ArrayExpress id: E-MTAB-4377).
We hypothesized that inter-individual variability in gene expression could be exploited to identify co-expressed genes in the human retina. We used the Spearman Correlation Coefficient (SCC) to quantify the co-expression of all the gene-pairs across the 50 samples. We then selected a threshold for the SCC of 0.85 (which corresponds to about 1.4% of all the possible gene-pairs) and included in the gene network only gene-pairs whose SCC was above 0.85. The resulting gene network consisted of 11,022 genes and 1,401,990 edges connecting gene-pairs.
The website includes the following sections: Network, Search, Browser and Annotation:
Network
The Network tab provides graphically represents the genes that are co-expressed according to the Spearman's Correlation Coefficient. Nodes represent genes while edges represent a significant correlation. It's possible to search and select a node by the Official Gene Symbol.
| Legend: |
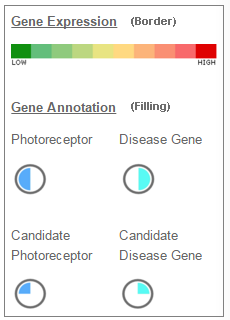
|
It is also possible to n save the network shown in the browser as an image or a table (.xls).
Search
In Search tabs (Search by Gene, Search by Position) it is possible to search by the Official Gene Symbol, by chromosome or by genomic coordinates. Results are shown in a tabular format.
Browser
A UCSC genome browser track reports the Average Coverage (AC) across the genomic sequence by counting the number of mapped reads per 100bp averaged across the 50 samples. The final length of genome that was covered by mapped reads was 238 Mb, corresponding to 7.78% of the genome.
Annotation
In Annotation tab a user-provided mutation list (VCF-file up to 2 Mb) can be uploaded. Each mutation is automatically annotated with the properties of the associated gene, i.e. expression level, transcript id, candidate disease gene and candidate photoreceptor gene.
How to cite
Michele Pinelli, Annamaria Carissimo, Luisa Cutillo, Ching-Hung Lai, Margherita Mutarelli, Maria Nicoletta Moretti, Marwah Veer Singh, Marianthi Karali, Diego Carrella, Mariateresa Pizzo, Francesco Russo, Stefano Ferrari, Diego Ponzin, Claudia Angelini, Sandro Banfi, Diego di Bernardo. An atlas of gene expression and gene co-regulation in the human retina. Nucl. Acids Res. (2016) doi: 10.1093/nar/gkw486. Abstract
Ruiz-Ceja, K.A., Capasso, D., Pinelli, M. et al. Definition of the transcriptional units of inherited retinal disease genes by meta-analysis of human retinal transcriptome data. BMC Genomics 24, 206 (2023). https://doi.org/10.1186/s12864-023-09300-w. Abstract
Privacy | Terms of Use | © 2015 Tigem | Designed by Diego Carrella
